Grevy’s Zebra Conservation Technologies
Integrating Genomes, Microbiomes, Parasites, Diets, and Demography.
Conserving large charismatic megafauna such as the Grevy’s zebra presents many challenges because numbers are low and populations are small and widely scattered. New technologies can change the scale and scope of the research and monitoring necessary for sustaining such species. This project was conceived and co-funded with Princeton University and the Mpala Research Centre in Nanyuki, Kenya. The San Diego Zoo’s Conservation Technology Department, which I created, was the largest benefactor, and the arrangement was formalized in a Memorandum of Understanding between all parties.
There were five primary research subprojects, outlined below.
with D. Rubenstein (Princeton), J. Ayroles (Princeton), S. Kocher (Princeton), D. Martins (Mpala, Kenya), T. Berger-Wolf (Ohio State), J. Holmberg (WildMe).
-

Drones and Machine Learning
Drone imagery and Artificial Intelligence (AI) offer new possibilities for mapping and classifying large tracts of lands on regular bases. High resolution images can detect long-term changes associated with different degrees of herbivore exclusion over a 5-year period. Longer term research will reveal the importance of various land types to Grevy’s zebra.
Above are shown a drone image (L) and the image classified by a supervised machine learning algorithm (R) into landscape categories.
-

Improved Immune Assays
New measurement technologies allowed quantification of immune responses from zebra fecal samples for the first time. This provided important insights into how immune responses to the same gastrointestinal parasites differ between species. Immunoglobin A (IgA) is the most prevalent antibody in the gut (where parasitic nematodes live), and helps reduce nematode condition and egg output. Grevy’s have the highest IgA levels among equids in this ecosystem.
-

Population Structure
We assessed population genetics of Grevy’s zebra using restriction site associated DNA (RAD) markers. RAD sequencing was run with 3074 single nucleotide polymorphisms (SNPs) on 17 tissue samples collected from 6 locations. Exploratory analyses suggested that the two population groups (Laikipia and Samburu) were not genetically distinct. Although a principle components analysis (left) showed mild separation between the two populations, this trend was not sustained in the neighbor-joining tree (right), where individuals from the two sites interspersed.
-

Upgrading Zebra Genomes
The only fully-assembled equid genome available today is the horse genome.
This severely limits our ability to use modern genomic techniques to study the life history and biology of these endangered and threatened species.
Using 10x genomics technologies, we have assembled the genomes for two zebra species: the Plains (Equus quagga) and Grevy’s (E. grevyi). Sequencing and assembly of the Mountain zebra (E. hartmannae) is in progress. Together, these three genomes will represent the best-assembled equid genomes outside of the horse, and provide an important resource for the population genomic work planned on Grevy’s populations.
-

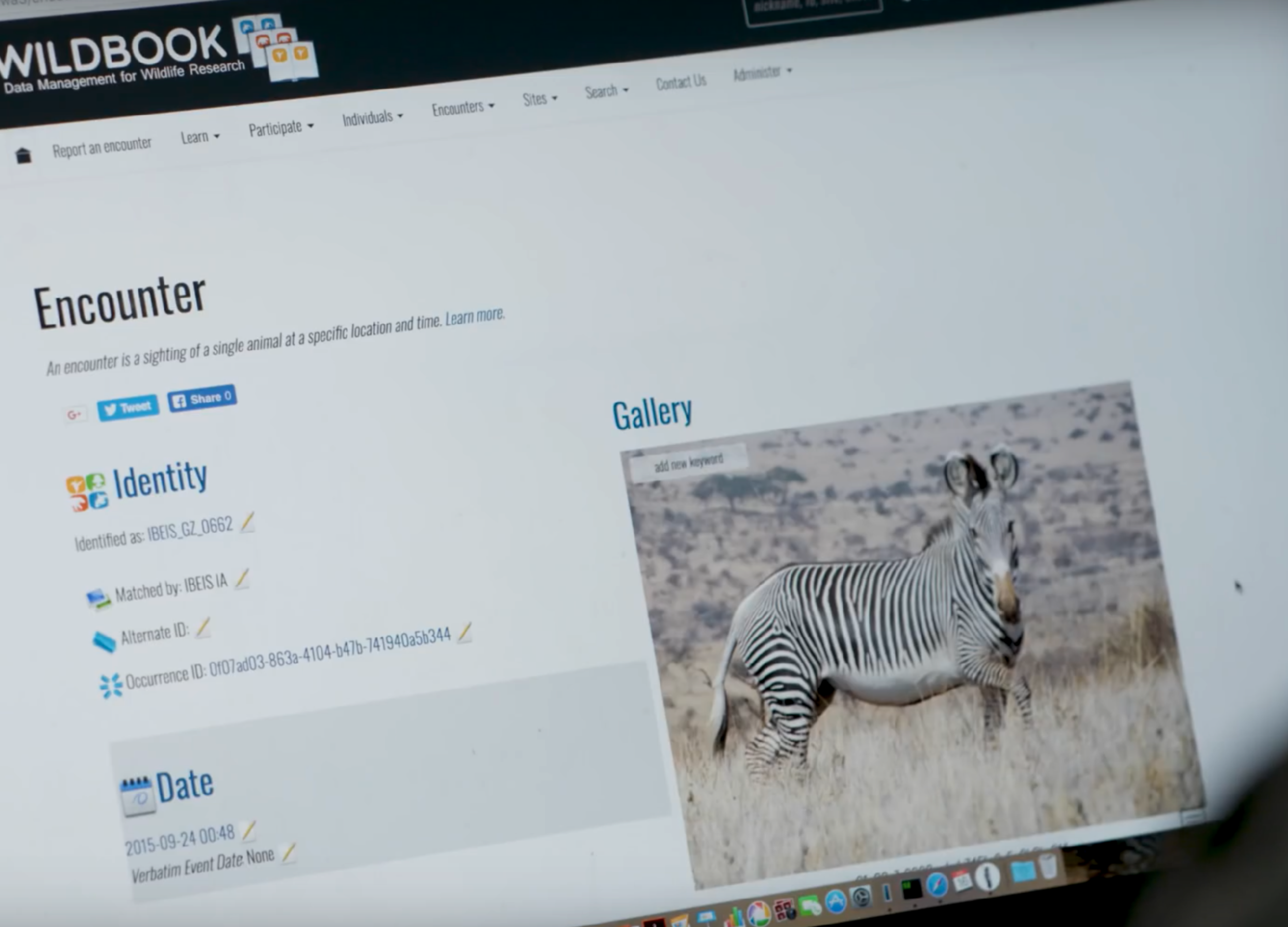
Individual Identification with Computer Vision
Using computer vision to recognize patterns on zebra bodies, we completed ecological surveys to estimate the endangered Grevy’s zebra population using mark-recapture techniques and hundreds of thousands of images.
These photographs were collected by hundreds of individuals, many of whom were volunteers, tourists, and students, in dozens of vehicles over a three-day period. Each camera is GPS-tagged.
This project developed the image-based ecological information system (IBEIS) used to track individual animals over time.
The resulting population estimates and distributions will inform future conservation and management initiatives.